Genetics
Homework Help & Tutoring
We offer an array of different online Genetics tutors, all of whom are advanced in their fields and highly qualified to instruct you.
Genetics
Before Mendel, the theory of inheritance stated that the parents' traits would blend to form the child's traits, meaning that a tall and short parent would produce a child of medium height. The majority of outcomes supported this idea. The theory failed spectacularly when two brown-eyed parents had a child with blue eyes. It was Mendel who developed a framework of heredity upon which the true nature of the gene would be illucidated. All students of genetics are familiar with the Punnett squares - the simple calculational tool used to determine the genotypes and phenotypes of offspring given the genes of the parents for a particular set of traits. If you have any doubts about the fact that all fields of knowledge are ultimately connected, including the fields of genetics and mathematics, take a look at the paper titled Game Theory Using Genetic Algorithms. The subject of game theory was discussed under math in the Subjects & Resources section of this website.
A good course in introductory genetics will involve study of the following topics:
- Genetics and the organism
- The Structure of genes and genomes
- Gene function
- The inheritance of genes
- Recombination of genes
- Gene interaction
- Gene mutations
- Chromosome mutations
- The Genetics of bacteria and phages
- Recombinant DNA technology
- Applications of recombinant DNA technology
- Genomics
- Transposable genetic elements
- Regulation of gene transcription
- Regulation of cell number: normal and cancer cells
- The genetic basis of development
- Population and evolutionary genetics
- Quantitative genetics
Students should follow the Journal of Genetics to stay up to date in this field. In addition, there is a Rutgers University genetics tutorial and a University of Utah genetics tutorial that are both worthy of a student's time. Good books on genetics can be found at Amazon.com and Google.
... ... ...
SECRETS of SEX DETERMINATION
In multicellular animals, there is a considerable difference between primary sexual differentiation and secondary sexual differentiation where the former occurs in the gonads and the latter occurs in the secondary sexual characteristics. The organisms with both the male and female sex organs are called hermaphrodites or bisexuals or monoecious while those only male or female producing fertile gametes are dioecious. Intersex is those that are intermediate between males and females.
TYPES OF SEX DETERMINATION
Sex determination is done either on the basis of chromosome or environmental factors and involves:
- Somatic sex determination,
- Germ –line sex determination
- Dosage compensation
In environmental determination, certain stimuli like temperature can influence the transformation or development of a male or female where both has the same genotype. Germ line sex determination depends on the ‘germ line sex determining gene’ and somatic environment. Chromosomal dimorphism (XY or ZW) is frequently associated with marked differences in the gene content of the sex chromosomes. Heterogametic sex (XY in males and ZW in females) has a single dose of same gene in contrast to homogametic sex (XX in females and ZZ in males). Hence the fundamental aspect of dosage compensation is to restore the balance between autosomal and chromosomal gene products in males and females.
SEX DETERMINATION IN DROSOPHILA
Drosophila contain 4 pairs of chromosomes of which 3 pairs are autosomes (Chromosomes II, III and IV) and one pair of sex chromosome (XX or XY). Thus the genetic constitution is 6A + XX and 6A + XY (or 2A +XX & 2A +XY, where A = number of autosomes)
Experiment by Bridges in relation to sex determination in Drosophila
Calvin Bridges experimented with sex determination in Drosophila and his studies can be grouped into two parts: experiments with non-disjunction of X chromosome of female flies and later, the genetic analysis of the triploid females and its progenies. During meiosis I and II of anaphase, segregation of the paired chromosomes usually take place but occasionally if the segregation fails then the event is called non-disjunction. This results in the production of two abnormal gametes, one with an extra chromosome (n+1) and the other having one chromosome less (n-1); the aneuploidy zygotes produced by the fertilization of such gametes (2n +1) or (2n -1).
Bridges observed that flies produced by non-disjunction had normal 6 autosomes along with an X0 (zero indicating absence of one chromosome) or XXY (one excess chromosome). The XXY flies developed into normal females while the XO developed into sterile males. Male characters were not produced in XXY flies in spite of having the Y chromosome and neither did XO flies develop female characters. From this observation, Bridges concluded that in Drosophila, the Y chromosome does not bear any male determining factor but is necessary for fertility since XO males are sterile. In 1922 he worked on triploid females (3A +XXX) that received 2 X from the mother and one X from the father. These were fertile and were experimentally mated with normal diploid (2A +XY) producing the following results:
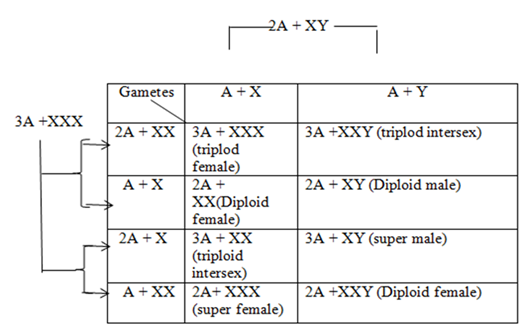
DEVELOPMENT OF GENIC BALANCE THEORY
Results indicated the Y chromosome doesn’t produce males in Drosophila. Instead the “male factor” is present in autosome and the female determining factor is located in the X chromosome and it was stated that males are produced due to absence of an X chromosome. This is the genic balance theory. Bridges proposed that when X: A ratio is 1:2 (X:2A) then it attains the threshold for maleness but the presence of one excess chromosome turns this balance towards female differentiation .
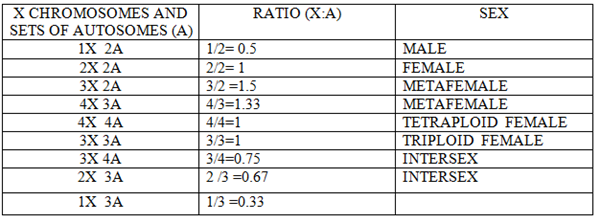
GENES IN SEX DETERMINATION
Discovery of genes controlling sex determination modified the genic balance theory and there are 4 genes in Drosophila that control sex determination:
a) sxl –sex lethal gene
b) tra and tra 2 (transformer genes recessive, autosomal)
c) dsx-double sex lethal gene on 3rd chromosome.
ROLES OF THE DIFFERENT GENES IN SEX DETERMINATION
Sxl is the most important gene in sex determination and its activity depends on the influence of some numerator genes on the X chromosome like sis a, sis b, sis c, and runt. The sxl group act as a master switch and the activity of the sxl is essential for female development. In the 2A + XX zygote having the ratio value of 1.0 acts as a signal for activation of sxl and sxl mRNA undergoes splicing to form functional Sxl protein. This Sxl protein can bind to tra mRNA and control its splicing to form normal Tra protein. Tra protein joins with Tra2 protein forming a Tra/Tra2 dimer. This dimer joins with the mRNA of dsx gene and controls its splicing so that the female determining Dsx protein is formed, which activates the sex determination genes that produce female specific proteins, hence a female fly is produced. ON the contrary, in XY or XO flies, (ratio 0.5), Sxl protein is not produced and accordingly active Tra proteins is also not formed. Under such conditions, Tra2 protein associates with dsx mRNA for splicing that produces male determining protein. Dsx male specific proteins are produced that activate the dosage compensation genes msl1, msl2, mle, mof and the male fly is produced. Sxl protein in (X:A=1.0) can produce female specific protein either by splicing regulation or transcriptional regulation of msl (male specific lethal) genes. Thus the genes for female development are sxl, tra, tra2, dsx and ix and those for dosage compensation in males due to absence of sxl are msl1, msl2, mle, etc.
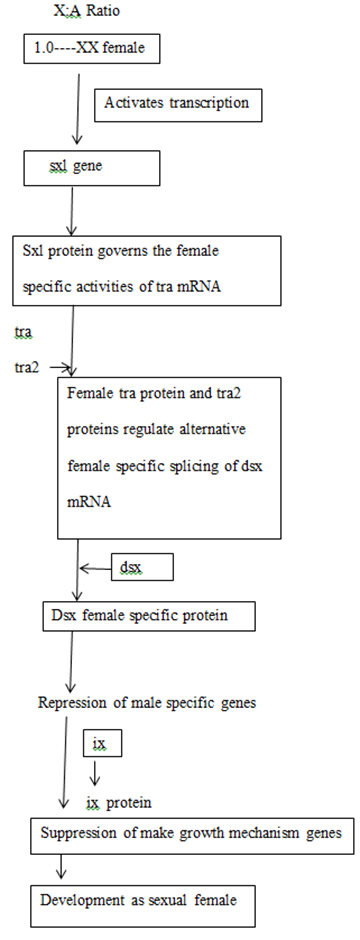
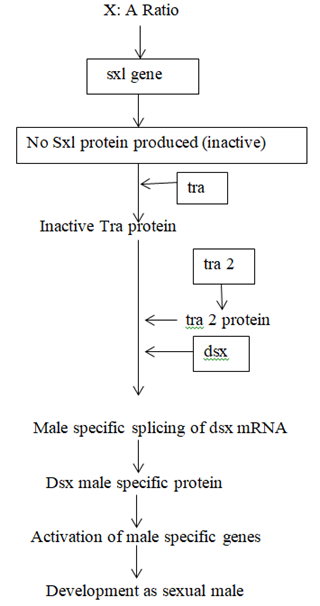
GENETIC REGULATION OF SEX DETERMINATION IN DROSOPHILA
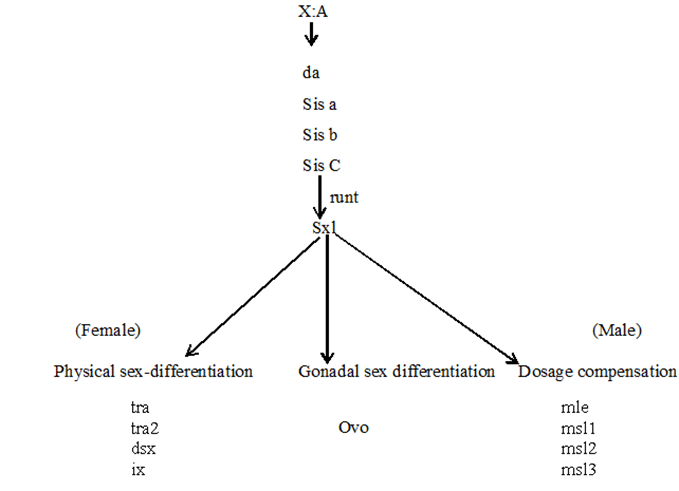
Relationship between sex determining genes in Drosophila
... ... ...
HOW TO TACKLE PROBLEMS ON GENE MAPPING
Six point checklist to solve gene mapping problems
Students pursuing genetics often come across problems on gene mapping, which refers to the process of mapping the loci of the genes on the chromosome with the distance expressed in Centimorgan. It actually is the percentage of the recombination between two loci considered as the map distance. In other words, the frequencies of recombinant gametes produced are used as an index of the distance between two loci on a chromosome as proposed by Sturtevant (while still an undergraduate!!). Hence 1 (one) Centimorgan is the map distance that equals 1% recombination between two loci.
It was Mendel in 1865 who proposed the transfer of “factors” from parent to the offspring as the process of transfer of traits from one generation to another; but it was not until 1900 that his work was rediscovered independently by Franz Correns, Erich Tschermak and Hugo de Vries who credited Mendel as the Father of Genetics. Eventually, the progress in research, and theories like Chromosomal theory of inheritance and the discovery of Linkage and Recombination by Morgan led to the development of new concepts of genes and chromosomes.
It was postulated that the movement of chromosomes was parallel to the movement of genes from one generation to another. The concepts of linkage and recombination led to the concept of linear location of genes on the chromosome. For the beginner, the initial question that may come to the mind is crossing over itself and its relationship to linkage. Crossing over or the process of recombination is actually the exchange of chromatid fragments between the non-sister chromatids of the homologous chromosome occurring during the pachytene stage of the first meiotic prophase. The point of crossing over is called the chiasma and the total number of chiasma on a chromosome is called chiasma frequency, which usually ranges from 1 to 12. The following process of gene mapping is based on three points crossing over.
-
Find out the number of parental and recombinant gametes.
In order to tackle the problem associated with gene mapping, the first thing that we need to find out is the exact number of gametes that can be produced from a cross. In general, the formula for the number of gametes produced in a cross is given by 2n where ‘n’ represents the number of alleles.
For example if the number of alleles is 2, the total number of gametes produced is 22 or 4. Similarly for the consideration of three alleles in a three point cross, the possible gametes produced is 23 or 8 in number
If we consider two linked alleles A and B distributed in a genotype A B / a b, then the types of gametes produced can be:
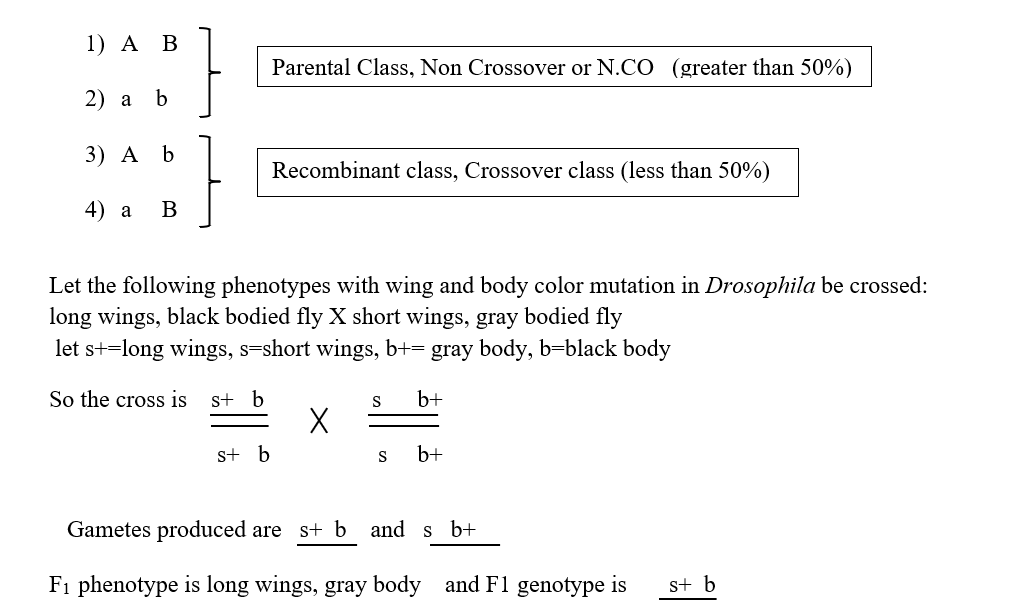
So now if a test cross (Cross with a double recessive parent) is performed with F1 female flies, the progeny types and proportions produced are parental combination 82% and recombinant combination 18%.
It is very important to remember that frequency of crossing over in meiotic tetrads is not the same as with the frequency of crossover or recombinant chromosomes. Linkage map distances are determined by the frequencies of crossover or recombinant meiotic crossover or recombinant chromosomes. Each meiotic crossing over event produces two crossover chromosomes, hence in a single crossover occuring between two loci in 100 % of the tetrads, only 50% of the produced chromosomes will be recombinant, so the recombination frequency will be 50%
Relative Frequency.
Relative frequency (RF), = (Number of recombinants / number of offsprings ) X 100
If 49 gametes are produced in meiosis without crossing over and only 12 are produced with crossover, then RF= (12 /100) x 100=12%
Three point crosses:
Let us consider a parental progeny with the following genotype
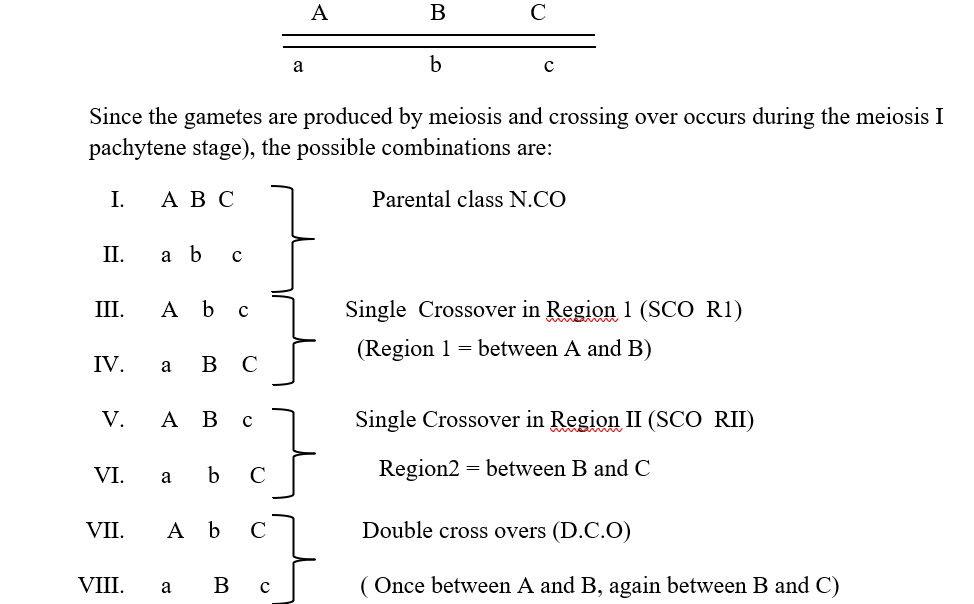
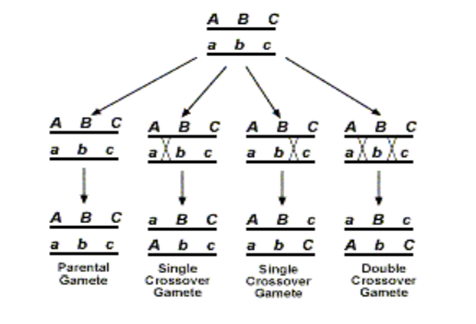
Out of the produced gametes, it is obvious that the parental class or NCO class gametes will occur at the highest frequency. This will be followed by the SCORI and SCO R II having intermediate frequencies of occurrence and this will be followed by the DCO or the double crossover class where the process of crossing over has occurred in both the region 1 as well as the region II together. Now since one crossover usually prevents another adjoining crossover from occurring, so the possibility of a double crossover between three alleles is very low and it will occur in the lowest percentage. It is also important to understands that map distances are additive. For example, if A and B are linked and are 5 map units apart, and loci A and C are linked by 3 map units ,then loci B and C are also linked and is either 2 map units or 8 map units apart (mu=map units)
Total = 1000
-
Selection of the DCO classes from the given problem
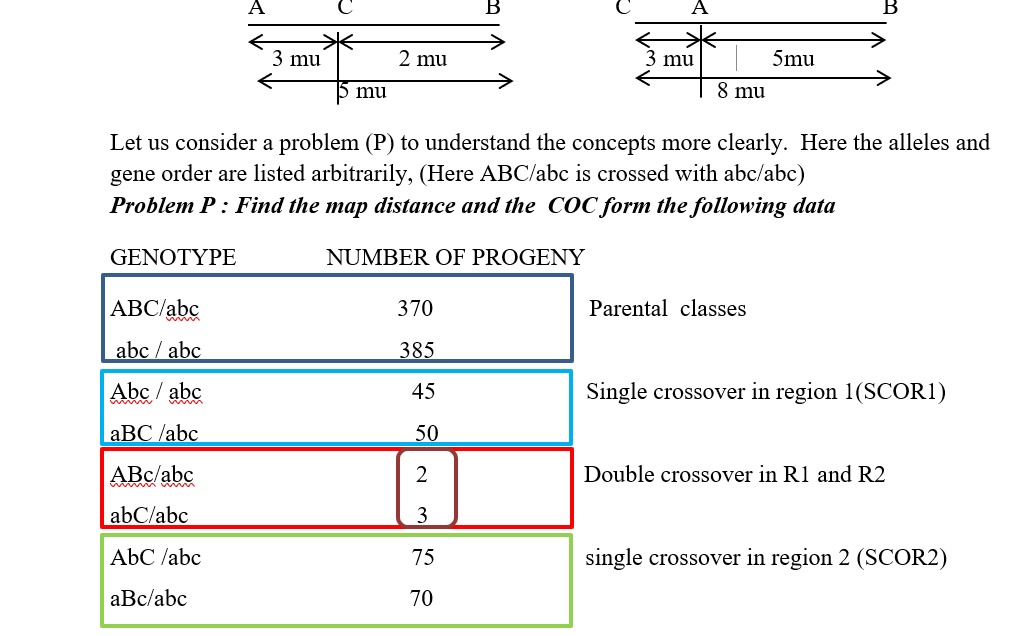
The most important thing you need to do is to find out the DCO class from the analysis of the given data. For that we should check whether all the 8 classes of progeny have been given or not. As you will remember, the probability of two successive crossing overs between three linked alleles in very low, so the best way to identify the DCO class is to look for the lowest number of progeny that has been produced. Hence from the given sample, the progeny with numbers 2 and 3 are the lowest, hence corresponding to the DCO class. Thus the DCO class is ABc / abc and abC/abc .Hence the genotype of the parent producing the DCO is ABc /abC.
In certain problems, the DCO class may be absent in the data implying the DCO is 0, even if one DCO class is given, the other should be calculated out.
-
Determining the central gene and then the gene order of other classes
In order to determine the gene order, we need to understand that only the central gene is inverted in the case of a DCO. We can approach the problem by considering each gene as the central gene and see if it tallies with the data.
Considering gene A is in the middle, we get BAC/bac as the parental genotype, so the DCO would be BaC /bAc, which does not tally with the DCO data. Hence A cannot be in the middle.
#considering B as the central gene, we get the DCO class as AbC /aBc. But this does not tally with the data.
#So now considering C as the central gene, we get DCO a AcB/ aCb. (Written as ABc and abC). These tally with the DCO data. Hence the gene order is A-C-B
Alternately, we can consider that the parental chromosomes are ABC and abc and the DCO progeny has the genotype of ABc and abC. The allele C is present with A and B on the parental chromosome and also with a and b on the DCO chromosome (and vice versa for c). Hence C must be at the center. So rearranging the gene order we get:
the actual NCO as ACB/acb
Hence SCORI is Acb/aCB
SCO R2 is ACb/acB and the DCO is AbC /aBc
-
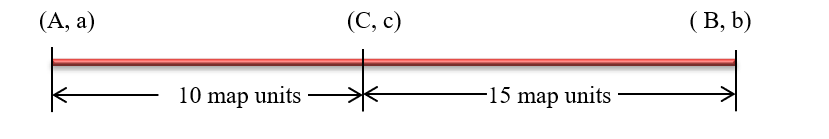
Calculating map distance
Now the order of the genes are known, the genetic map can be completed by calculating the linkage distances between the loci A and C and C and B .The parental marker combinations for the A-C interval are A C and a c; the recombinants are Ac and aC.
The formula for calculating map distances between Ac intervals is the map distance and is given by:
Map distance = (All SCO RI +all DCO) / Total number of progeny X 100
Hence the distance between A--C is (45+50 + 2 +3)100 X 100 = 10%
Similarly, the map distance between C—B is (75+70 +2+3)/1000 x 100=15%
Note that the progeny with the genotype resulting from the double cross overs are included in the numerator of the equation since one of the two crossovers occurred within the A-C interval. Again, consider that progeny with the genotypes resulting from double crossovers resulting in the C-B interval (Cb and cB) are also included in the numerator as before.
Thus the map distance between A –B is 25 map units or centimorgan (as map distances are additive).
5. Creating a genetic map
In creating the genetic map, you must be careful to remember the gene order and clearly draw the map. The loci of the alleles should be pointed with a vertical line and distances marked appropriately to nullify confusion.
-
Interference and Coincidence of crossing over (COC)
If we consider the given problem P, if the F1 trihybrid is considered there are 5 progenies resulting from the double-cross. So what is the number of the observed double crossover gametes? It is 2 +3 =5, so out of 1000 total progeny, only 5 are double crossover types, so is 0.5% or its frequency is .005. Now if we consider that the process of double crossover is completely random and is a result of two independent cross overs occurring together, then the expected frequency of the double crossover class should be obtainable from the map distances between A-C-B.
The expected frequency of Double cross over should be 0.10 x 0 .15(10 and 15 map units ) or 0.015. So why was this frequency not attained? The reason for this is that fewer double crossovers are prevented by chromosomal interference or chiasma interference. One crossover decreases the probability of another crossover. The degree of coefficient of coincidence (COC) is given by:
COC = observed frequency of DCO/ expected frequency of DCO
Hence in problem P, the COC = .005/.015 = 0.33.
In the absence of any interference, COC=1. Sometimes the coefficient of interference is given by the formula 1- COC. Hence the Coefficient of interference in problem P is 1 – 0.33= 0.67.
By now the concept and method of working out map distances should be clear for you but you must practice multiple times to gain mastery. Best of luck!
To fulfill our tutoring mission of online education, our college homework help and online tutoring centers are standing by 24/7, ready to assist college students who need homework help with all aspects of genetics. Our biology tutors can help with all your projects, large or small, and we challenge you to find better genetics tutoring anywhere.
College Genetics Homework Help
Since we have tutors in all Genetics related topics, we can provide a range of different services. Our online Genetics tutors will:
- Provide specific insight for homework assignments.
- Review broad conceptual ideas and chapters.
- Simplify complex topics into digestible pieces of information.
- Answer any Genetics related questions.
- Tailor instruction to fit your style of learning.
With these capabilities, our college Genetics tutors will give you the tools you need to gain a comprehensive knowledge of Genetics you can use in future courses.
24HourAnswers Online Genetics Tutors
Our tutors are just as dedicated to your success in class as you are, so they are available around the clock to assist you with questions, homework, exam preparation and any Genetics related assignments you need extra help completing.
In addition to gaining access to highly qualified tutors, you'll also strengthen your confidence level in the classroom when you work with us. This newfound confidence will allow you to apply your Genetics knowledge in future courses and keep your education progressing smoothly.
Because our college Genetics tutors are fully remote, seeking their help is easy. Rather than spend valuable time trying to find a local Genetics tutor you can trust, just call on our tutors whenever you need them without any conflicting schedules getting in the way.